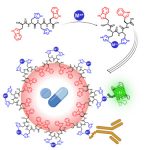
Our work in the area of nanoscale structures focuses on the use of our approach for understanding the relationship between physical properties and structure. This work has recently focussed more specifically in the development and application of methods to discover new soft materials (e.g., gels, emulsions, etc.).
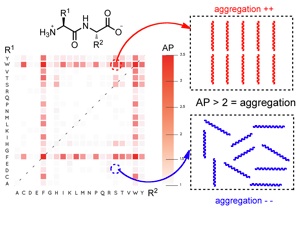
Initial work in this area focussed on the type of methods that were able to predict the ability of small molecules to self-assemble and act as low molecular weight delators. Our initial work in this area of predicting the self-assembly propensity of molecules focused on the use of coarse grained methods to determine the self-assembly of dipeptides. This work led to the development of a new metric the peptide’s Aggregation Propensity (AP score) as a convenient method for scoring a peptide’s ability to aggregate and as such to self-assemble.

Following the successful demonstration of the methodology and the AP score to act as a predictor for the ability of a peptide to assemble, we applied this methodology to discover the first examples of uncapped, tripeptide hydrogelators. This work resulted in the development of the hydrophobicity corrected AP score.
Our work in this area has developed rapidly to include the development of a new class of emulsifiers (via nano fibrous networks) and functionalism gels through the process of cooperative self-assembly (co-assembly). In addition to the development and application side of our work in this area, we also maintain a strong focus on understanding the physical principles that control the properties of these materials.
The experimental validation of the computational predictions is made possible through a longstanding collaboration with our excellent colleagues in the Ulijn Lab at the Advanced Science Research Centre (ASRC) attached City University New York.
Recent Structure Publications
-
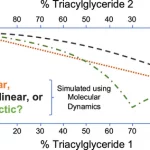
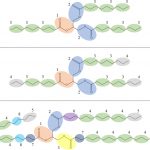 Compr. Rev. Food Sci. Food Saf. 2024, 23, e13316.
Compr. Rev. Food Sci. Food Saf. 2024, 23, e13316. -
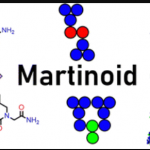 Phys. Chem. Chem. Phys. 2024, 26, 4939.
Phys. Chem. Chem. Phys. 2024, 26, 4939. -
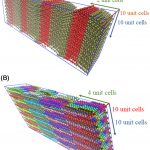 J. Comput. Chem., 2023, 44, 1795.
J. Comput. Chem., 2023, 44, 1795.